Disease Markers
Aging and Disease biomarkers continue to be a significant part of metabolomic research. Prognostic markers, disease progression markers that reflect disease pathways are ever increasing often combined with clinical data and other analytes such as proteins and RNA expression. At HMT, our OMEGA Scan advanced and LC-OMEGA Scan advanced provide over 1000 metabolites found in blood, plasma and serum to cover the biodiversity in human conditions and allow for new metabolite identifications. All our animal and human study analysis starts with sample quality assessments.
Step One: Review project plans. Source of samples, sample tracking. Balanced groups. Availability of clinical data.
Step Two: Decide on right platforms: OMEGA Scan (Polar metabolites LC-OMEGA (lipid metabolites MSCAN (Lysophosolipids, sphingomyelins, lipid mediators). In addition to untargeted analysis, a panel of 353 metabolites can be quantified in plasma, serum, urine, stool or any biological sample. Using two or three platforms, over 1000 metabolites are annotated providing an extensive pool for biomarker discovery. Quantitative data (110 or 353 metabolites) can be used for more exact ratio calculations and for quality analysis. HMT has a virtual healthy human plasma data base (over 900 metabolites) that can be used to establish quality of controls and discover biomarkers from metabolites that are out of range.
Step Three: Quality analysis: In plasma samples is there evidence of thermal decomposition, hemolysis, platelet contamination or long term storage markers that may influence analysis. Statistical analysis includes looking for disease and treatment subtypes.
Step Four: Drawing biological, pathway and disease markers requires deep probing into subtypes and response markers combined with clinical data.
Vaccines
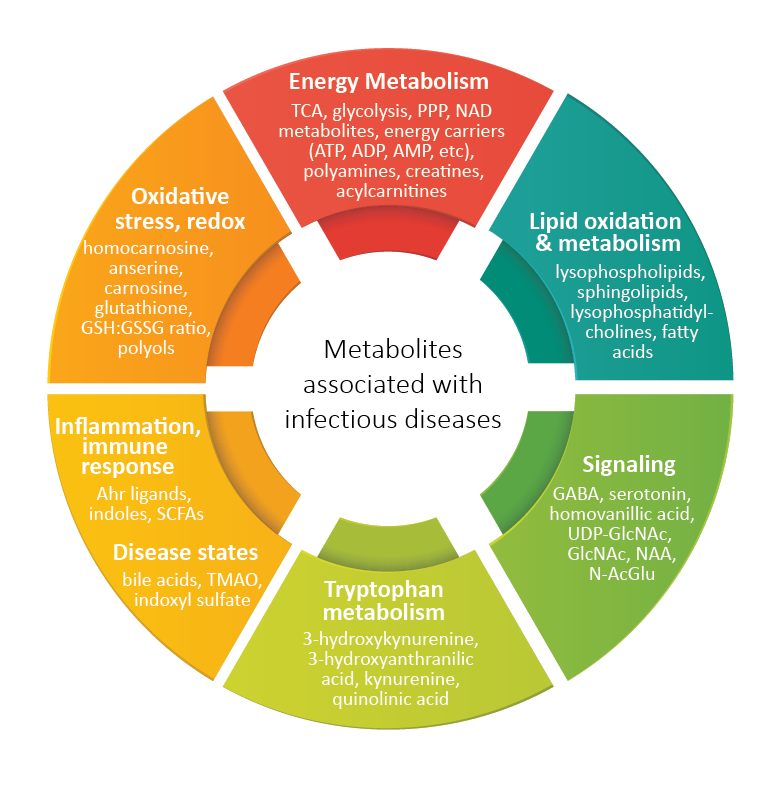 As the world continues to recover and learn from the devastating effects of the COVID-19 pandemic, there is an increasingly urgent demand for vaccines and new therapeutics to combat the virus. HMT offers collaborative support to scientists and clinicians working to develop these drugs and vaccines. With our metabolomics technology, we can help to identify biomarkers and novel drug targets, as well as time-dependent metabolic changes and characteristics that are able to predict the drug and vaccine effectiveness and/or toxicity. Here, we propose some ideas on how metabolomics can be extremely useful for pre-clinical and clinical studies.
As the world continues to recover and learn from the devastating effects of the COVID-19 pandemic, there is an increasingly urgent demand for vaccines and new therapeutics to combat the virus. HMT offers collaborative support to scientists and clinicians working to develop these drugs and vaccines. With our metabolomics technology, we can help to identify biomarkers and novel drug targets, as well as time-dependent metabolic changes and characteristics that are able to predict the drug and vaccine effectiveness and/or toxicity. Here, we propose some ideas on how metabolomics can be extremely useful for pre-clinical and clinical studies.
HMT caught up with Dr Chan, the lead author of the paper “Metabolic perturbations and cellular stress underpin susceptibility to symptomatic live-attenuated yellow fever infection”, which includes key findings based on HMT’s metabolome analysis and was published in top biomedical journal Nature Medicine last August. Dr. Chan’s team focused on the differences in blood profiles of symptomatic and asymptomatic individuals before and after inoculation with live yellow fever viral vaccine. Transcriptome analysis and metabolome analysis performed on blood specimens revealed that individuals with endoplasmic reticulum (ER) stress and reduced tricarboxylic acid (TCA) cycle activity have increased susceptibility to symptomatic outcomes after inoculation with the vaccine.
HMT had the opportunity to ask Dr. Chan via e-mail about examples of metabolome analysis in his research, and the significance and prospects of using metabolomics in infectious diseases research. Read his interview HERE
Application Examples
- Metabolic perturbations and cellular stress underpin susceptibility to symptomatic live-attenuated yellow fever infection
- Prevalence of Slow-Growth Vancomycin Nonsusceptibility in Methicillin-Resistant Staphylococcus aureus
- Prominent Steatosis with Hypermetabolism of the Cell Line Permissive for Years of Infection with Hepatitis C Virus
- Hepatitis C Virus Infection Promotes Hepatic Gluconeogenesis through an NS5A-Mediated, FoxO1-Dependent Pathway
